

New achievements for banana streak virus diagnostics, TropAg
November 8, 2019
The Crawford Fund’s Queensland Committee has again partnered with TropAg2019 to assist 10 young researchers from developing countries attend and present their science at this international conference which will be held in Brisbane from 11-13 November 2019.
Successful candidates were chosen by a selection panel made up of representatives of The Crawford Fund and the TropAg2019 conference organisers, based on submitted abstracts of their research.
In the lead-up to the conference we will be publishing short blog posts written by the young researchers about their work. Here is the eighth blog.
By Thu Ha Ngo, University of Queensland
Banana streak virus (BSV) remains the most challenging of all banana viruses to detect because of their broad genetic and serological diversity, as well as the fact that some banana cultivars have both endogenous and exogenous forms of viral DNA.
The presence of endogenous BSV causes false positive results for conventional PCR, which means that this assay cannot be used for diagnosis of infection. Most diagnostic assays for BSV rely on serological methods which depend on the limited sources of antigens and antibodies worldwide. The BSV capsid protein contains an N-terminal, intrinsically disordered (NID) domain that is surface-exposed on the virion and likely is multifunctional and plays important roles in viral replication and transmission. The immunodominant continuous epitopes on the virion are also located in the NID domain, and therefore this domain is of great interest from a diagnostics perspective.
Using chemically synthesized peptides to mimic the continuous epitopes of five BSV species including Banana streak Mysore virus (BSMYV), Banana streak Obino I’Ewai virus (BSOLV), Banana streak Imove’ virus (BSIMV), Banana streak Goldfinger virus (BSGFV) and Banana streak Cavendish virus (BSCAV), antisera have been raised in rabbits, and shown specificity to each virus species and sensitivity in a range of assay formats such as Western Blot, enzyme-linked immunosorbent assay (ELISA), immunosorbent electron microscopy and immunocapture PCR. For BSMYV, antibodies were produced against synthetic peptide E1 with carrier protein KLH, E-BSA mixture of epitope E1 and E2 with carrier protein BSA, and E-MAP8 is the dendrimer construct of epitope E1 and E2 without carrier protein. Antibodies against E-MAP8 showed the best sensitive reaction. For other four BSV species, the dendrimer construct MAP8 of epitope was applied to produce polyclonal antibodies. This work confirmed the applicability of synthesis epitopes/peptides to produce antipeptide antibodies for BSV diagnostics when the native protein is absent.
The Oxford MinION Nanopore sequencing system is a promising new technology for point-of-care diagnostics. We have successfully used this system and generated long reads with greater than 99% accuracy, allowing accurate and sensitive detection of BSV. The long reads allow episomal from endogenous sequences to be differentiated.
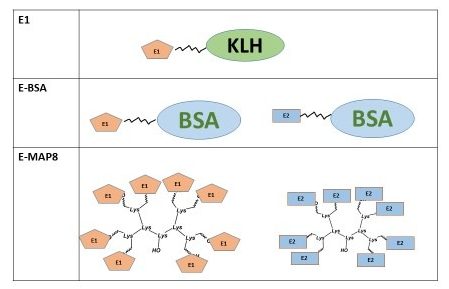
Figure 1a. Structure of synthetic peptides for Banana streak Mysore virus (BSMYV) E1 with carrier protein KLH; E-BSA mixture of epitope E1 and E2 with carrier protein BSA; and E-MAP8 is the dendrimer construct of epitope E1 and E2 with no carrier protein.
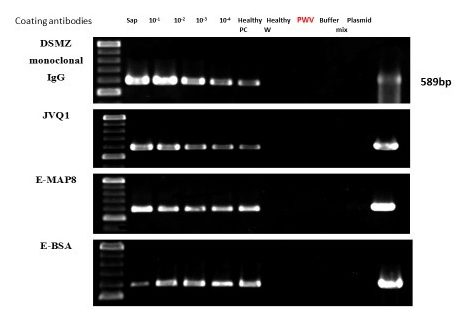
Figure 1b. Immunocapture PCR results for ten-fold dilution series of plant sap to examine the sensitivities of antipeptide antibodies against BSMYV. Concentration of antibodies in the first dilution was 5 µg/ml. 589 bp amplicon was observed on the gel using all antibodies against BSMYV to capture antibodies. DSMZ commercial antibodies and JVQ1 polyclonal antibodies were used in this study as positive control. Negative control PWV IgG was used to coat the tube, and no amplification was observed, demonstrated that PWV IgG didn’t bind to the BSMYV virion in sap extract. The two healthy controls including cv. Pisang Ceylan (AAB genome) and cv. William (AAA genome) were negative on the gel. IC PCR was able to detect BSMYV at 1:10000 dilution of sap extract.




